Documentation
Tutorial
1. Input a target
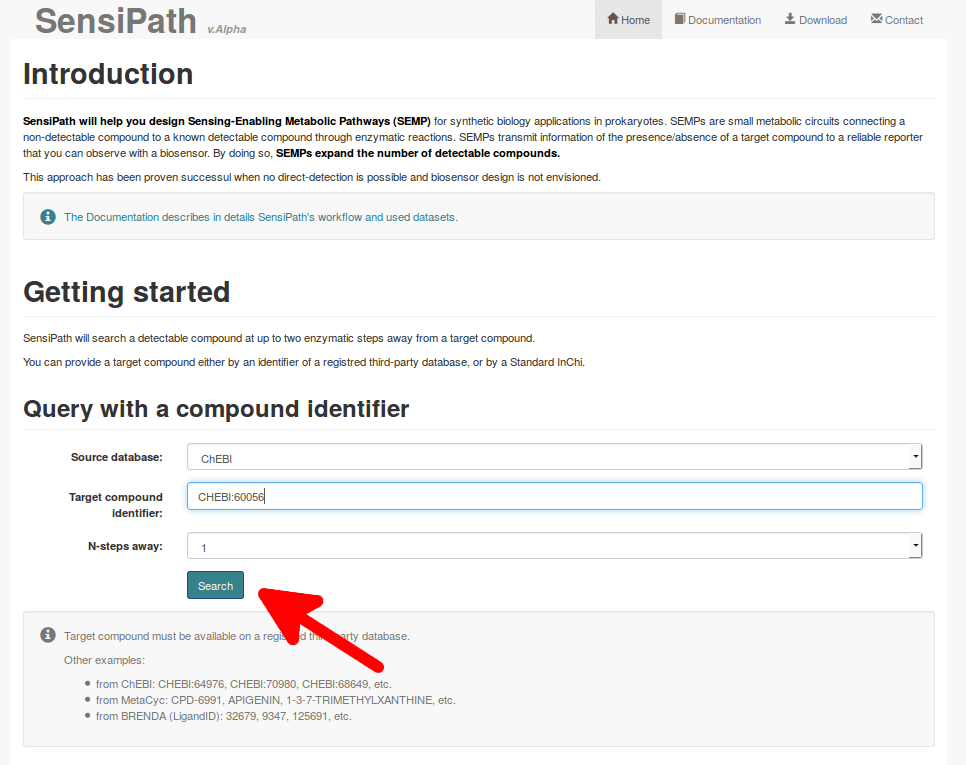
Input as target the compound that you wish to detect within the cell. You can input your favorite target as an external database identifier (e.g. CHEBI:27732 from ChEBI) or as a Standard InChI ("IUPAC Chemical Identifier"). Standard InChI are easy to retrieve from chemical compounds databases (such as PubChem), or you can use tools to draw and convert your target into Standard InChI (PubChem Sketcher for instance).
"N-steps" parameter sets the maximum number of reactions in the Sensing-enabling Metabolic Pathways (SEMPs).
We recommend the use of identifiers from ChEBI and to restrict the query to one-step SEMPs.
2. Wait patiently...

Once the target is recognized, SensiPath will process it through all our collected enzymatic reactions. This computation can take a long time and depends directly on the number of reactions able to transform the target into another compound. Note that if you choose to process more than one enzymatic step ("N-steps" parameter), products from the previous step will become themselve targets at the next iteration step. Hence, a longer waiting time might be expected for higher number of steps.
For easy retrieval of results, you can bookmark this page and retrieve your results later.
3. Browse the results
Results are displayed on three views:
- Informations: a summary of your query, results and tips to go further;
- Pathways: a list of Sensing-enabling Metabolic Pathways, linking your target to a known detectable compound through enzymatic reactions;
- Graph: an interactive metabolic graph displaying all reached compounds.
You can access the different views by the tabs at the top of the page.
3.1 Informations
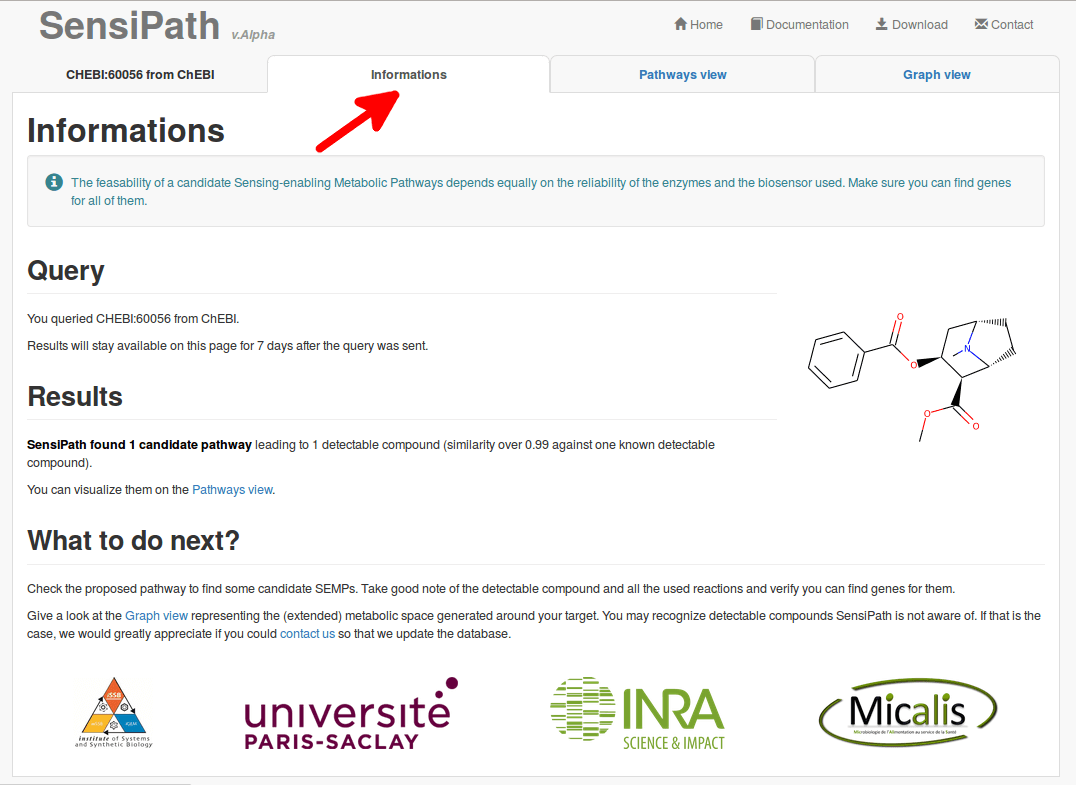
3.2 Pathways view

The list is ordered by putative detectable compound. Each arrow represents a set of biochemical reactions which are identical as far as SensiPath is concerned. In the same way, compounds' graphical representation (and their name) shows one example from a set of compounds which are internally considered identicals. Please read the documentation about compounds and reaction encoding to know more about this feature.
SensiPath aggregates several information sources to compute the pathways. Sometimes databases can contain information that is not accurate. It is responsability of the user to check in the literature relevant informations about all biochemical reactions and detectable compounds in order to assess the viability of the design. Moreover, if you wish to do in vivo experiments, you should also check that all needed genes are suitable to work in your chassis.
SensiPath provides cross-references to all its sources. You can click on arrows (reactions) and compounds to display that information.
3.3 Graph view
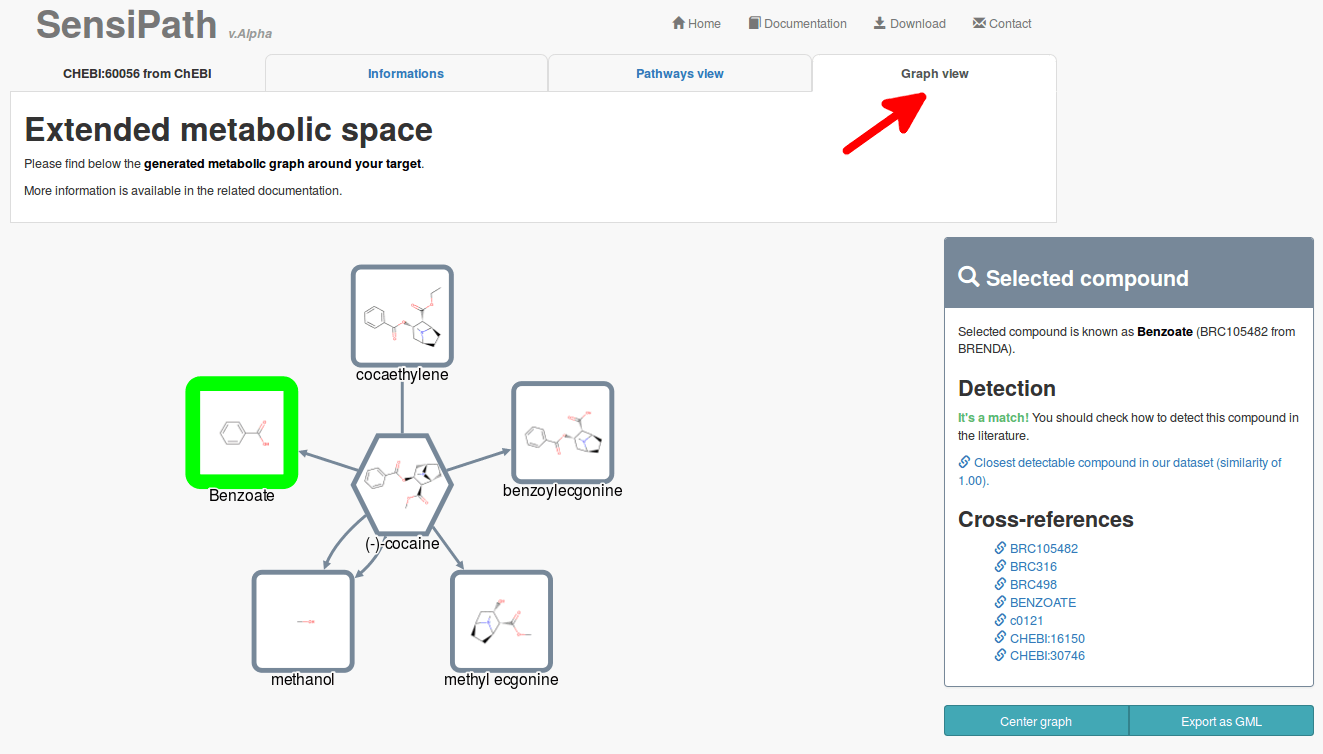
The graph displays all reachable compounds from your target. Putative detectable compounds are depicted with green border. The intensity of the green depends on its best similarity score against our collected list of detectable compounds.
This view should be used to assist you further in the design if you cannot find interesting data on the Pathway view. Graph view can help you to find alternative solutions if you are aware of additional detectable compounds not yet included in our dataset.
Both reactions (edges) and compounds (nodes) are clickable and will display information on the right panel. You can download the graph as a Graph Markup Language file (GML) that you can read and modify with network visualization tools (such as Cytoscape).
FAQ
What is SensiPath? What is a Sensing-Enabling Metabolic Pathway?
SensiPath is a web-server assisting the design of Sensing-Enabling Metabolic Pathway (SEMPs). SEMPs create a link between a non-detectable target compound and a known detectable compound through enzymatic reactions, so that the information of the presence/quantity of the target is transferred to the detectable compound.
Such pathways can be used to design sensing circuits for metabolic engineering or synthetic biology applications.
Do I need SensiPath? What are the applications which benefit from this strategy?
If you need to assess the intracellular presence/quantity of a compound for your application (e.g. to design a whole-cell biosensor, to perform high-throughput screening, dynamic regulation, etc.), but cannot find nor design a direct-sensing solution, then SensiPath could help you. Of course, if you already have a direct-sensing solution such as a riboswitch or an allosteric transcription factor for your target compound, then you should try out those solutions in priority.
How does it work?
The overall idea is that the information about the presence/absence of a non-detectable target compound can be transferred through enzymatic reactions to a compound for which a biosensor exists.
Users provide their favorite target compound, then SensiPath tries all the known biochemical reactions on this compound using a special reaction encoding. Thus, “reachable” compounds from the target are retrieved in a so-called metabolic graph. Known detectable compounds can be mapped into this graph, and the pathways linking the target to the detectable compounds can be extracted. See the technical section for more details.
Does it really work?
Detecting compounds through the metabolism is a powerful approach which is quite easy to try in vivo. SensiPath’s algorithms and method have been used thoughtfully in our group and others, which led to the design of several new whole-cell biosensors.
For more information, please see:
- Libis, V., Delépine, B., Faulon, J.L. (2016) Expanding biosensing abilities through computer-assisted design of metabolic pathways. ACS Synth. Biol. doi:10.1021/acssynbio.5b00225
- Rogers, Jameson K., and George M. Church. Genetically Encoded Sensors Enable Real-Time Observation of Metabolite Production. Proceedings of the National Academy of Sciences, February 8, 2016, 201600375. doi:10.1073/pnas.1600375113.
- Silva-Rocha,R. and de Lorenzo,V. (2014) Engineering Multicellular Logic in Bacteria with Metabolic Wires. ACS Synth. Biol., 3, 204–209. doi:10.1021/sb400064y
- Xue,H., Shi,H., Yu,Z., He,S., Liu,S., Hou,Y., Pan,X., Wang,H., Zheng,P., Cui,C., et al. (2014) Design, construction, and characterization of a set of biosensors for aromatic compounds. ACS Synth Biol, 3, 1011–1014. doi:10.1021/sb500023f
What are SensiPath’s limits?
SensiPath rely on external databases as information sources for biochemical reactions, metabolites and known detectable compounds. Those dataset are far from being complete or error-proof. Subsequently, SensiPath will propagate the limitations of those databases.
Moreover, Sensing-Enabling Metabolic Pathways can suffer from the same limits as any heterologous pathway, such as intermediate compound toxicity, gene expression difficulties, etc.
Last but not least, it is sometimes tedious to find a sequence for an annotated enzyme (or a transcription factor). As a rule of thumbs, we recommend our users to always search in the literature for a working example in their chassis for both the enzymes and the intracellular biosensor before starting their project.
How to use SensiPath?
Please see the Tutorial on this page, or try out the examples.
How to cite SensiPath?
If you found SensiPath useful and wish to mention our web service, please cite:
- Delépine,B., Libis,V., Carbonell,P. and Faulon,J.-L. (2016) SensiPath: computer-aided design of sensing-enabling metabolic pathways. Nucl. Acids Res., doi:10.1093/nar/gkw305.
- Libis, V., Delépine, B., Faulon, J.L. (2016) Expanding biosensing abilities through computer-assisted design of metabolic pathways. ACS Synth. Biol. doi:10.1021/acssynbio.5b00225.
Biology-related questions
For which chassis is designed SensiPath?
SensiPath was designed with its use for prokaryotes in mind (especially E.coli), but the biochemical reactions were collected from any organism.
What if my target compound cannot enter the cell?
Sensipath currently predicts sensing pathways involving intracellular biosensors. Depending on the host, the cell membrane might not be permeable to the target compound. If a compound does not diffuse passively or through transporters it will not be processed by neither the metabolic nor sensing module and detection might fail. Sensipath does not predict the permeability aspect for a given compound/host and therefore searching for such information in the literature is advised prior in vivo implementation. Several strategies may circumvent permeability issues. One possibility is to switch to in vitro biosensing through the use of cell-free extracts lacking membrane. Alternatively, if downstream intermediates of the SEMP can go through the membrane more easily than the target compound, secretion or extracellular display of the metabolic module might be an interesting strategy.
What about target specificity?
A specific limitation of SEMPs resides in their inability to discriminate between the presence in the medium of the target molecule or of any of the intermediates of the SEMP. In the context of whole cell biosensors, this can be easily circumvented by the use of control strains lacking enzymatic steps to allow discrimination.
Another specificity issue might emerge from the fact that both transcription factors and enzymes can display high level of ligands promiscuity. This feature has been tuned by evolution in a particular context and varies from gene to gene. If specificity of the sensor is important for a given application, it is advised to search in the literature for gene variants showing reduced promiscuity. It is important to note that an important degree of promiscuity of several elements composing a SEMP (enzymes, transcription factors) does not necessarily translate into poor sensor discrimination abilities. Indeed, chances for the promiscuous abilities to be cumulative in terms of specificity decrease in a synthetic heterologous pathway are rather low. Discrimination issues emerge either if the first step of the pathway can transform several input compounds into the same output metabolite transmitted to the downstream steps of the SEMP, or if alternative output metabolites can serve as effectors to the transcription factor. Importantly, promiscuity has been subjected to directed evolution experiments with a lot of success.
Can I trust SEMPs using highly abundant compounds?
Some compounds (such as cofactors) are highly abundant in cells, and it may seem a priori difficult to detect variations of their availability. Sensors for such compounds are natural regulators of cell metabolism. Therefore, there is a growing interest in the community in taking advantage of such “generic sensors” for abundant chemicals because, under suitable conditions, could potentially be re-used for many biotech applications. To our knowledge, this originated from the findings of Siedler et al. 2014 and Knudsen et al. 2014 and their use of a NAD(P)H sensor to select for enzyme specificity through high-throughput screening (with special experimental conditions).
Thus, its seems nowadays that even highly abundant chemicals could be suitable for sensing.
What about detection range?
Characteristics of a SEMP such as the limit of detection, the dynamic range (i.e. the difference between the un-induced and fully induced output signal) and the linear range of detection depend mainly on the characteristics of the transcription factor that is used.
In our experimental validations the SEMPs behaved in the exact same way as the transcription factor would do in presence of its natural inducer.
Limiting enzymatic steps might cause interference in the transfer of the signal in some case but this was not observed in the test cases we choose.
Technical questions
What is SensiPath's version?
SensiPath is available in version v2.1.2 since June 2017.
Where does SensiPath's data come from?
Biochemical reactions:
- MetaCyc (v19.1)
- Rhea (v66)
- BRENDA (v15.2)
- EAWAG-BDD2 (accessed in Decembre 2015)
Detectable compounds:
- RegTransBase (v7)
- RegPrecise (v4)
- RegulonDB (v90)
- BioNemo (v6.0)
How are normalized and encoded compounds?
Normalization is necessary so that similar compounds are matched together with our encoding. Compounds are notably normalized by striping salts, removing all charges and all hydrogens. Compounds without explicit structural information are ignored. Stereochemistry is conserved.
Compounds are encoded with an ECFP-like fingerprint called molecular signature ("Stereo Signature Molecular Descriptor"). You can download the software to generate molecular signature from SourceForge.
Molecular signatures define a compound as a list of overlapping circular topological fragments. Each fragment (called atomic signature, aka atomic environment) is a canonized representation of the direct neighbourhood of an atom, in term of atom-type, bond-type and stereochemistry. Molecular signature's most important parameter is its diameter. It determines the area covered by each fragment.
For instance, below is the molecular signature of caffeine (CHEBI:27732) at diameter 2:
2.0 [O](=[C]) 3.0 [N]([C][C][C]) 1.0 [N]([C]=[C]) 1.0 [C]([N][N]=[O]) 1.0 [C]([N]=[N]) 3.0 [C]([N]) 1.0 [C]([C][N]=[O]) 1.0 [C]([C]=[C][N]) 1.0 [C](=[C][N][N])
Caffeine at diameter 12:
1.0 [O](=[C]([N]([C][C](=[C,1]([N]([C][C,2]))[N](=[C,2])))[N]([C][C]([C,1]()=[O])))) 1.0 [O](=[C]([C](\=/[C]([N,1]([C])[N](=[C,2]))[N]([C][C,2]))[N]([C][C]([N,1]()=[O])))) 1.0 [N]([C][C](\=/[C]([C,1](=[O])[N]([C][C,2]))[N](=[C,2]))[C]([N]([C][C,1])=[O])) 1.0 [N]([C][C]([C]([N]([C][C,1](=[O]))=[O])\=/[C]([N,2][N]([C][C,1])))[C](=[N,2])) 1.0 [N]([C][C]([C](=[C,1]([N](=[C,2]))[N]([C][C,2]))=[O])[C]([N]([C][C,1])=[O])) 1.0 [N]([C](\=/[C]([C]([N,1]([C])=[O])[N,2])[N]([C][C]([N,1]()=[O])))\=/[C]([N,2]([C]))) 1.0 [C](\=/[C]([C]([N,1]([C])=[O])[N]([C][C,2]))[N]([C][C]([N,1]()=[O]))[N](=[C,2])) 1.0 [C]([N]([C][C]([C]([N]([C][C,1])=[O])=[C,2]))\=/[N]([C,2]([N]([C][C,1](=[O]))))) 1.0 [C]([N]([C][C](=[C,1]([N]([C][C,2]))[N](=[C,2])))[N]([C][C]([C,1]()=[O]))=[O]) 1.0 [C]([N]([C](\=/[C]([C,1](=[O])[N]([C][C,2]))[N](=[C,2]))[C]([N]([C][C,1])=[O]))) 1.0 [C]([N]([C]([C]([N]([C][C,1](=[O]))=[O])\=/[C]([N,2][N]([C][C,1])))[C](=[N,2]))) 1.0 [C]([N]([C]([C](=[C,1]([N](=[C,2]))[N]([C][C,2]))=[O])[C]([N]([C][C,1])=[O]))) 1.0 [C]([C](\=/[C]([N,1]([C])[N](=[C,2]))[N]([C][C,2]))[N]([C][C]([N,1]()=[O]))=[O]) 1.0 [C]([C]([N]([C][C,1](=[O]))=[O])\=/[C]([N]([C][C,1])[N](=[C,2]))[N]([C][C,2]))
Each line correspond to an atomic signature (with its occurrence).
In current implementation of SensiPath, we use a diameter of 12 to encode all compounds.
For more information, please see:
- Carbonell, P., Carlsson, L., Faulon, J.L. Stereo signature molecular descriptor. Journal of Chemical Information and Modeling, 53(4): 887-897, 2013. doi:10.1021/ci300584r
How are encoded reactions?
Reactions are encoded with reaction signatures (σ represents molecular signature): \[\sum_\ \sigma_{products} - \sum_\ \sigma_{substrates}\]
Note that reaction signatures are centered around the reaction center. Subsequently, other atomic signatures are ignored. It allows us to have a fuzzy definition of reactions (depending on the diameter of the molecular signatures). This is considered a feature and is used to model enzymatic promiscuity.
Stereochemistry is inherited from molecular signatures. Reactions involving compounds without structural data are ignored.
This approach was notably used for the retrosynthetic tool XTMS.
For more information, please see:
- Carbonell, P., Carlsson, L., Faulon, J.L. Stereo signature molecular descriptor. Journal of Chemical Information and Modeling, 53(4): 887-897, 2013. doi:10.1021/ci300584r
- Carbonell, P., Parutto, P., Baudier, C., Junot, C., Faulon, J.L. Retropath: automated pipeline for embedded metabolic circuits. ACS Synthetic Biology, 3 (8), 565-577, 2014. doi:10.1021/sb4001273
- Carbonell, P., Parutto, P., Herisson, J., Pandit, S., Faulon, J.L. XTMS: Pathway design in an eXTended Metabolic Space. Nucleic Acids Research, 42 (W1): W389-W394, 2014. doi:10.1093/nar/gku362
What is enzymatic promiscuity?
Enzymatic promiscuity concept states than some enzymes are able to process several structurally similar substrates. Wikipedia has a more detailed definition of enzymatic promiscuity.What is an (extended) metabolic graph? The (extended) metabolic space?
A metabolic graph is a representation of a network of metabolic reactions (edges) and compounds (metabolites, nodes). We define the metabolic space as a theoric "whole" metabolic graph, having all known compounds and all known biochemical reactions.
For instance, Sensipath's "graph view" shows a metabolic graph centered on your target compound; it represents the compounds reachable at up-to two steps around the target in the metabolic space.
We use the term "extended" whenever we assumed enzymatic promiscuity to generate a metabolic graph. This is always the case with reaction signatures and depends on the diameter.
What are the “cross-references”? How are compounds/reactions matched together?
For both compounds and reactions, cross-references are links to external databases. All the elements under the same “cross-references” section have the same encoding (same signature) and are considered identical as far as SensiPath is concerned.
You will see that compounds (or reactions) that are matched together are sometime different from a biological point of view. This is expected and due to the normalization steps we used to import the compounds in our database. You can think of it as a fuzzy search.
How are computed compound similarities?
We use python library RDKit (release 2015.3.1) to compute similarities. All similarities are Tanimoto performed on compounds represented by RDKit’s ECFP4 fingerprint implementation.
A Tanimoto of 1 is a perfect match. We consider a "match" whenever two compounds have a Tanimoto over 0.99.
Troubleshooting
Error message: "Unfortunately, an error has occured. [...] is not a known atomic signature.”
The compound you provided has fragments (“atomic signatures”) which does not match with any compound in our database. Subsequently, even if we could apply some reactions on your compound, we could not retrieved their products or predict if they are detectable.
You can contact us to propose a new database to include in SensiPath, or to discuss other solutions for your project.
Results without sensing-enabling metabolic pathways
If no detectable compound is reachable around the target, then no sensing enabling metabolic pathway can be generated and the Pathway view is disabled.
Results without graphical representation of the metabolic graph
If there are too many reachable compounds around your target, the graph visualization will not be comfortable enough to be efficiently used. To prevent such issue, SensiPath will not display the metabolic graph.
If you queried at two-steps, you may try at one-step. Indeed, if a cofactor (or another highly connected compound) has been found at one step, SensiPath will try to display all its reactions at two-steps, which can lead to a very high number of compounds to display.
Note that even if the graph is not displayed, you can always download it as a GML for offline usage with a graph visualization software (such as Cytoscape).
Target compound is alone on the graph representation
Unfortunately, SensiPath was unable to find any biochemical reaction for your target, and thus cannot find pathways. It does not mean your compound cannot be processed by any enzyme, just that we are not aware of it.
This may be caused by the way we import reactions in SensiPath. For instance, if no structural information can be found (or it is incomplete, with poorly defined R-groups), the reaction is dropped.
If you feel like SensiPath should have found something, do not hesitate to contact us.