Introduction
SensiPath will help you design Sensing-Enabling Metabolic Pathways (SEMP) for synthetic biology applications in prokaryotes. SEMPs are metabolic circuits connecting a non-detectable compound to a known detectable compound through enzymatic reactions. SEMPs transmit information of the presence/absence of a target compound to a reliable reporter that can be integrated into a biosensor. By doing so, SEMPs expand the number of detectable compounds.
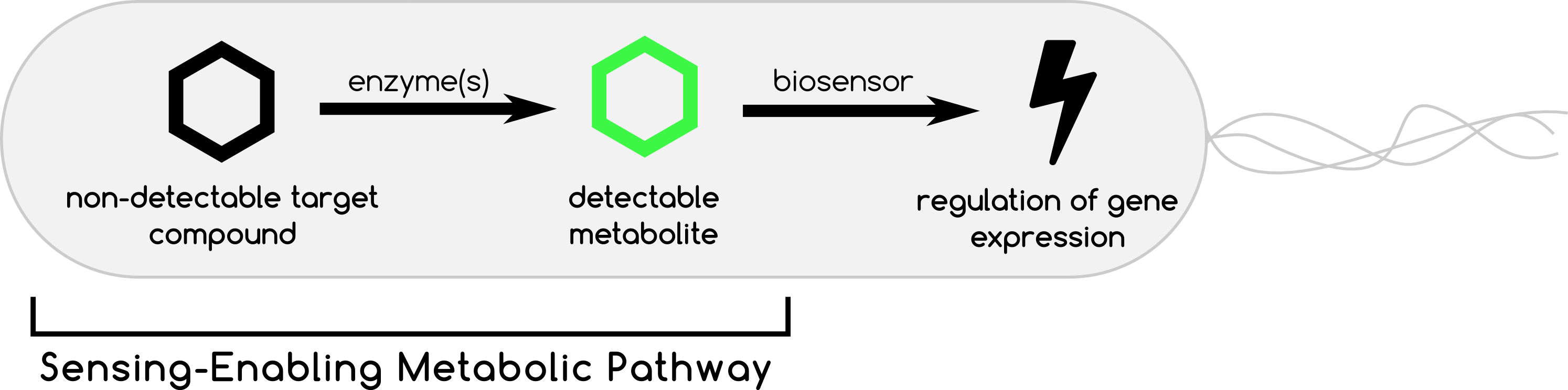
This approach has been proven successful when no direct detection is possible and as an alternative to biosensor design.
Getting started
SensiPath will search for a detectable compound up to two enzymatic steps away from any given target compound.
You can provide a target compound either by an identifier of a registered third-party database, or by a Standard InChI.
Please have a look at our tutorial if you seek more explanations about query fields, or just go ahead and try sample data below.
Query with a Standard InChI
Target compound represented by its InChI will be converted in molecular signature and matched in the database for querying.
If you want to quickly and easily convert a compound to a Standard InChI, we would recommend using PubChem Sketcher.